If you use NumPy for numerical computations, which I recommend if you program in Python, one of the key things to do is to process entire arrays in one go as much as possible (this is also true for MATLAB). Using these so-called vectorized operations makes sure that the operation is run using compiled code instead of interpreted Python code, since the array and matrix operations behind NumPy (and MATLAB) are implemented in compiled languages such as C.
As an example, I’ll create a Python program that produces the classical Mandelbrot fractal. This fractal is computed by defining, for each \(c\in\mathbb{C}\), a polynomial \(f_c(z)=z^2+c\). This polynomial is then iterated starting with \(z=0\), that is, \(f_c^n(0)\) is computed, and any point \(c\) for which \(f_c^n(0)\) does not “escape to infinity” for \(n\to\infty\) is part of the Mandelbrot set (there are some example values of \(c\) in the article Mandelbrot Set).
In order to compute this polynomial for all pixels of an image at the same time, we create matrices \({\bf Z}\) and \({\bf C}\), and then do the computation
\[{\bf Z}^{(t+1)}={\bf Z}^{(t)}*\,{\bf Z}^{(t)}+\,{\bf C},\]
where \(*\) means element-wise matrix multiplication. The matrix \({\bf Z}^{(0)}\) must be initialized to all zeros, so that’s easy.
The Matrix C
Each element of the matrix \({\bf C}\) must be initialized to the coordinate of the pixel in the complex plane that it represents, i.e., the pixel \((x,y)\) must get the value \(x+yi\). This makes sure that the correct function \(f_c\) is computed at each point. A straightforward way to do that is to create a row of x coordinates and a column of y coordinates. For example, for the points with integer coordinates in the range \([-2,2]\), this would be
\[\begin{pmatrix}
-2 & -1 & 0 & 1 & 2
\end{pmatrix}\]
and
\[\begin{pmatrix}
2 \\
1 \\
0 \\
-1 \\
-2
\end{pmatrix}.\]
By replicating both to a full \(5\times5\) matrix and adding them, with the second one multiplied by \(i\), we get
\[\begin{pmatrix}
-2+2i & -1+2i & 2i & 1+2i & 2+2i \\
-2+i & -1+i & i & 1+i & 2+i \\
-2 & -1 & 0 & 1 & 2 \\
-2-i & -1-i & -i & 1-i & 2-i \\
-2-2i & -1-2i & -2i & 1-2i & 2-2i
\end{pmatrix}.\]
In practice, a basic plot of the Mandelbrot fractal can be made on a grid that extends from \(-2\) to \(1\) in direction of the x-axis, and \(-1\) to \(1\) in the direction of the y-axis. The magnitude of the elements of a \(480\times320\) matrix with this range is shown in Figure 1.
 Figure 1. Magnitude of C.
Figure 1. Magnitude of C.One way to compute \({\bf C}\) in Python is the following.
import numpy as np m = 480 n = 320 x = np.linspace(-2, 1, num=m).reshape((1, m)) y = np.linspace(-1, 1, num=n).reshape((n, 1)) C = np.tile(x, (n, 1)) + 1j * np.tile(y, (1, m))
This code literally translates the vector and matrix operations from the example above. There are alternatives such as mgrid() that could be used, but I think that the construction using tile() explicitly is clearer in this case.
Iterating the Polynomial
We now have the matrices \({\bf Z}\) and \({\bf C}\) ready to start iterating. This is where it gets a bit tricky. The basic code is completely trivial:
Z = np.zeros((n, m), dtype=complex) for i in range(100): Z = Z * Z + C
However, there’s a problem. The values that “escape to infinity” grow so quickly that they overflow the maximum float value in no time, which first results in Inf values and then quickly in NaN values. To avoid this, I’ll add a mask M of pixels that are potentially in the Mandelbrot set, but from which we will remove pixels if we discover that they are not. Mathematically, it can be proven that pixels values with a magnitude larger than 2 will escape and cannot be part of the set. Hence, the code can be adapted as follows.
Z = np.zeros((n, m), dtype=complex) M = np.full((n, m), True, dtype=bool) for i in range(100): Z[M] = Z[M] * Z[M] + C[M] M[np.abs(Z) > 2] = False
There are two important constructs here. The first one is the notation Z[M]. This is so-called boolean indexing. It selects the elements of the matrix Z for which M contains True. This enables you to only continue iterating on those pixels that have not escaped yet. The second line updates M itself. After each iteration, we determine all pixels that have escaped, using the expression np.abs(Z) > 2. We then use boolean indexing again to remove these pixels from M, using the expression M[np.abs(Z) > 2] = False.
Hence, using array operations allows computing the Mandelbrot set in only a few lines of code, and with much better performance than by iterating over all pixels with Python for loops. Figure 2 shows the result of running the above code (so, for 100 iterations) and then plotting M.
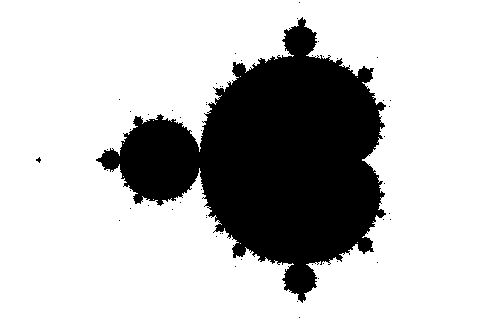 Figure 2. Approximation of the Mandelbrot set after 100 iterations.
Figure 2. Approximation of the Mandelbrot set after 100 iterations.This black-and-white image shows the Mandelbrot set in black. For more information on the difference between this and the usual colorful images that you possibly expected here, see again the article Mandelbrot Set.
Python Code
For completeness, a complete Python program follows.
import numpy as np from imageio import imwrite m = 480 n = 320 x = np.linspace(-2, 1, num=m).reshape((1, m)) y = np.linspace(-1, 1, num=n).reshape((n, 1)) C = np.tile(x, (n, 1)) + 1j * np.tile(y, (1, m)) Z = np.zeros((n, m), dtype=complex) M = np.full((n, m), True, dtype=bool) for i in range(100): Z[M] = Z[M] * Z[M] + C[M] M[np.abs(Z) > 2] = False imwrite('mandelbrot.png', np.uint8(np.flipud(1 - M) * 255))
The call to flipud() makes sure that the plot is not upside down due to the matrix indexing that NumPy uses. This does not make any difference for this image, since the Mandelbrot set is symmetrical around the x-axis, but it becomes important when you zoom in on different regions of the set.
Hi Tom!
In the second last line, you mean symmetric around x-axis, right?
I also have a doubt -
why the lower bulb in the Mandelbrot set looks bit tilted? (I went through other similar graphs over internet and everywhere is is like that)
Yes, around the x-axis, of course. Thanks, I've adapted the text. Both the upper and lower small bulbs are indeed tilted. This is not an effect of the way in which the image was produced or something, it is really how the Mandelbrot set looks.
Add new comment