This is part three of a tutorial that describes how to create a reconstruction of a cone-beam computed tomography (CT) dataset (also read part one, part two, and part four)). In this part, I’ll show how to convert the preprocessed images from part two into projections.
The term projection has a very specific meaning in the context of tomography, as explained in the first article of my introductory series of articles, Tomography, Part 1: Projections. For completeness, the following Section is a small recap of that material, but the main focus of this tutorial is the practical stuff that follows after that.
Recap
What is needed for tomographic reconstruction is that each projection image consists of line integrals through the object. I.e., the value of every pixel in the image should represent the sum of all the material of the object on the line between the X-ray source and the detector. The preprocessed images from part two of this tutorial, however, show the X-ray photons that remain after passing through the object. That is not a projection.
How to correct for this? Note that the solution is not simply inverting the image (i.e., mapping an image in the range \([0,1]\) to the range \([1,0]\) by setting the value of each pixel \(v\) to \(1-v\)). I’ve seen people do that, but it’s wrong. The following formula describes the relation between the intensity \(I(s)\) of the X-ray beam and the density \(\mu(s)\) of the object, which are both dependent on the distance \(s\) traveled through the object.
\[I(s)=I(0)\exp\left(-\int_0^s\!\mu(x)\,\mathrm{d}x\right)\]
The integral sums the density \(\mu\) along the path through the object, and the exponential models the fact that the photon absorption is not linear. \(I(0)\) is known, since it is the initial intensity of the X-rays. \(I(s)\) is what is measured at the detector. To single out the integral, we take the logarithm of both sides of the equation, to get
\[p(s)\equiv-\ln\left(\frac{I(s)}{I(0)}\right)=\int_0^s\!\mu(x)\,\mathrm{d}x.\]
This \(p(s)\) is a projection that represents the sum of the density along a line. So, to prepare X-ray images for tomographic reconstruction, take the ratio of photons that arrive at the detector and photons that were produced by the source, and take the negative logarithm of that.
In Practice
In an ideal world, your scanner would provide you with a perfectly calibrated source and detector. However, as was already clear from the need to have part two of this tutorial, this is not the case in practice. Given that we did the correction with the dark frames and the flat fields, however, what we can do is compute a single initial intensity and use that for all the pixels in a given image. Let’s call that \(I_0\).
How to estimate the \(I_0\) of a corrected X-ray image? Because there is always noise, we need to average many pixels to get a good estimate. In principle, all the pixels that contain no part of the object could be used (since no photons were absorbed by the object in the ray that hit that pixel), but you don’t want to adapt your algorithm for each object that you scan. A procedure that should work for many datasets, is to use the mean of two narrow strips of pixels at each side of the detector (Figure 1 shows the regions that I’ve used for my dataset). Given that you should really avoid that the object cuts through the side of the detector in your X-ray images, this should be applicable in general. The dataset that I use in this tutorial was made by a competent guy (thanks, Jelle! ☺), so this is okay in my case. For your own datasets, you should check all the images, of course.
 Figure 1. Pixels used to estimate initial intensity.
Figure 1. Pixels used to estimate initial intensity.I compute a separate \(I_0\) for each corrected X-ray image. Using \({\bf X}\) again as a matrix representing a complete corrected X-ray image, the projection image \({\bf P}\) can be computed as
\[{\bf P}=-\ln\left(\frac{\bf X}{I_0}\right),\]
with \(I_0\) a scalar and where the logarithm is taken pixel wise. Figure 2 shows the projection that corresponds to the image of Figure 1.
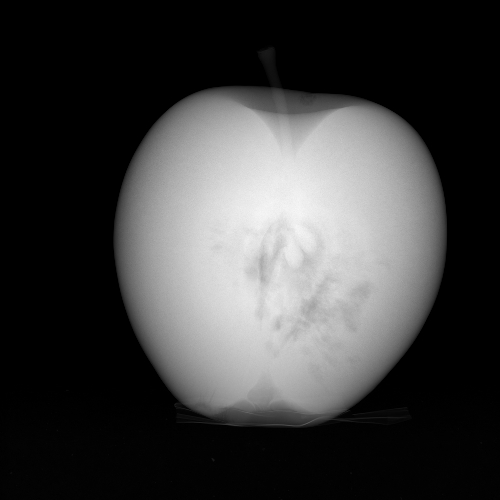 Figure 2. Projection image.
Figure 2. Projection image.Python Code
Below is a complete Python program that does all the preprocessing from the raw X-ray images all the way to the final projection images, as described in part one, part two, and the current article. A few remarks follow.
- This script imports the file
config.pyfrom part one. - I interpolate between the before and the after flat fields. This is not described in the text, but it will improve the reconstruction later.
- Before taking the negative logarithm, all pixels with a value higher than \(I_0\) are set to \(I_0\). This happens because of noise, and it corresponds with unphysical negative densities. An alternative is to replace all negative values by zero after taking the negative logarithm.
- The script also saves the preprocessed files before converting them to projections. This is for illustration only, you won’t need these for the reconstruction.
- Because I need the maximum over all images to create nicely scaled TIFF files, I do the processing twice. This is not an example of great coding, but it works for all dataset sizes (and tomography datasets can be huge). A more efficient option, cpu-wise, is to keep all the images in memory (in a 3D array), compute the maximum, and then save all the images. This works only if the complete dataset fits inside the memory of your computer, of course. Another alternative is to directly save the NumPy arrays in in floating point format, so that you don’t need the maximum. But I did what I did because TIFF files are easier to view, and because there are now no constraints on the memory of your computer anymore.
#filename: preprocess.py from __future__ import print_function from __future__ import division from os import makedirs from os.path import join, exists import numpy as np from imageio import imread, get_writer import config as cfg def save_tiff(name, im_in): # Expects image as floating point in range [0, 1]. im = im_in.copy() im = np.round(im * 65535).astype(np.uint16) with get_writer(name) as writer: writer.append_data(im, {'compress': 0}) if not exists(cfg.preproc_dir): makedirs(cfg.preproc_dir) if not exists(cfg.proj_dir): makedirs(cfg.proj_dir) # Dark frame. dark_frame = \ imread(join(cfg.raw_dir, 'dark_frame.tif')).astype(float) # Flat fields. pre_flat_field = \ imread(join(cfg.raw_dir, 'pre_flat_field.tif')).astype(float) pre_flat_field -= dark_frame post_flat_field = \ imread(join(cfg.raw_dir, 'post_flat_field.tif')).astype(float) post_flat_field -= dark_frame num_proj = cfg.num_projections # Determine maximum of projections. M = -np.inf # max for proj in range(num_proj): print('Preprocessing image {} (step 1)'.format(proj)) im = imread(join(cfg.raw_dir, 'raw{:04d}.tif'.format(proj))).astype(float) im -= dark_frame # Compute interpolated flat field. flat_field = \ ((num_proj - 1 - proj) * pre_flat_field + proj * post_flat_field) / \ (num_proj - 1) im /= flat_field I0 = np.mean([np.mean(im[:, : cfg.margin]), np.mean(im[:, -cfg.margin :])]) # Values above I0 are due to noise and wil produce negative densities later. im[im > I0] = I0 im = -np.log(im / I0) if np.max(im) > M: M = np.max(im) # Convert raw images to projections. for proj in range(num_proj): print('Preprocessing image {} (step 2)'.format(proj)) im = imread(join(cfg.raw_dir, 'raw{:04d}.tif'.format(proj))).astype(float) im -= dark_frame # Compute interpolated flat field. flat_field = \ ((num_proj - 1 - proj) * pre_flat_field + proj * post_flat_field) / \ (num_proj - 1) im /= flat_field I0 = np.mean([np.mean(im[:, : cfg.margin]), np.mean(im[:, -cfg.margin :])]) # Values above I0 are due to noise and wil produce negative densities later. im[im > I0] = I0 save_tiff(join(cfg.preproc_dir, 'prep{:04d}.tif'.format(proj)), im / I0) im = -np.log(im / I0) im /= M save_tiff(join(cfg.proj_dir, 'proj{:04d}.tif'.format(proj)), im)
Part four of this tutorial finally shows how to create the actual reconstruction.
Hi! If I want to add Poisson noise to the data, do I add it on the raw X-ray images or the projection data? I keep reading from papers that it has to be added to the projection data but it doesn't look good at all in this apple dataset when I do that. Thanks!
I've also always applied :-) it to the projection images myself. There's an example in ASTRA Toolbox Tutorial: Reconstruction from Projection Images, Part 1. (You are aware that you'd normally not do that for an actual dataset like this one, of course.)
Add new comment